tsfresh.feature_selection package¶
Submodules¶
tsfresh.feature_selection.feature_selector module¶
Contains a feature selection method that evaluates the importance of the different extracted features. To do so, for every feature the influence on the target is evaluated by an univariate tests and the p-Value is calculated. The methods that calculate the p-values are called feature selectors.
Afterwards the Benjamini Hochberg procedure which is a multiple testing procedure decides which features to keep and which to cut off (solely based on the p-values).
-
tsfresh.feature_selection.feature_selector.benjamini_hochberg_test(df_pvalues, hypotheses_independent, fdr_level)[source]¶ This is an implementation of the benjamini hochberg procedure that calculates which of the hypotheses belonging to the different p-Values from df_p to reject. While doing so, this test controls the false discovery rate, which is the ratio of false rejections by all rejections:
![FDR = \mathbb{E} \left [ \frac{ |\text{false rejections}| }{ |\text{all rejections}|} \right]](../_images/math/40805bea42d6f4ef764dca8bf9e6b7356aa4b76a.png)
References
[1] Benjamini, Yoav and Yekutieli, Daniel (2001). The control of the false discovery rate in multiple testing under dependency. Annals of statistics, 1165–1188 Parameters: - df_pvalues (pandas.DataFrame) – This DataFrame should contain the p_values of the different hypotheses in a column named “p_values”.
- hypotheses_independent (bool) – Can the significance of the features be assumed to be independent? Normally, this should be set to False as the features are never independent (e.g. mean and median)
- fdr_level (float) – The FDR level that should be respected, this is the theoretical expected percentage of irrelevant features among all created features.
Returns: The same DataFrame as the input, but with an added boolean column “rejected”.
Return type:
-
tsfresh.feature_selection.feature_selector.check_fs_sig_bh(X, y, n_processes=2, chunksize=None, fdr_level=0.05, hypotheses_independent=False, test_for_binary_target_real_feature='mann')[source]¶ The wrapper function that calls the significance test functions in this package. In total, for each feature from the input pandas.DataFrame an univariate feature significance test is conducted. Those tests generate p values that are then evaluated by the Benjamini Hochberg procedure to decide which features to keep and which to delete.
We are testing
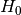 = the Feature is not relevant and can not be added
= the Feature is not relevant and can not be addedagainst
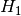 = the Feature is relevant and should be kept
= the Feature is relevant and should be keptor in other words
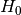 = Target and Feature are independent / the Feature has no influence on the target
= Target and Feature are independent / the Feature has no influence on the target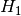 = Target and Feature are associated / dependent
= Target and Feature are associated / dependentWhen the target is binary this becomes
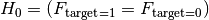
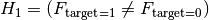
Where
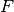 is the distribution of the target.
is the distribution of the target.In the same way we can state the hypothesis when the feature is binary
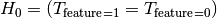
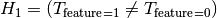
Here
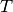 is the distribution of the target.
is the distribution of the target.TODO: And for real valued?
Parameters: - X (pandas.DataFrame) – The DataFrame containing all the features and the target
- y (pandas.Series) – The target vector
- test_for_binary_target_real_feature (str) – Which test to be used for binary target, real feature
- fdr_level (float) – The FDR level that should be respected, this is the theoretical expected percentage of irrelevant features among all created features.
- hypotheses_independent (bool) – Can the significance of the features be assumed to be independent? Normally, this should be set to False as the features are never independent (e.g. mean and median)
- n_processes (int) – Number of processes to use during the p-value calculation
- chunksize (int) – Size of the chunks submitted to the worker processes
Returns: A pandas.DataFrame with each column of the input DataFrame X as index with information on the significance of this particular feature. The DataFrame has the columns “Feature”, “type” (binary, real or const), “p_value” (the significance of this feature as a p-value, lower means more significant) “rejected” (if the Benjamini Hochberg procedure rejected this feature)
Return type:
tsfresh.feature_selection.selection module¶
This module contains the filtering process for the extracted features. The filtering procedure can also be used on other features that are not based on time series.
-
tsfresh.feature_selection.selection.select_features(X, y, test_for_binary_target_binary_feature='fisher', test_for_binary_target_real_feature='mann', test_for_real_target_binary_feature='mann', test_for_real_target_real_feature='kendall', fdr_level=0.05, hypotheses_independent=False, n_processes=2, chunksize=None)[source]¶ Check the significance of all features (columns) of feature matrix X and return a possibly reduced feature matrix only containing relevant features.
The feature matrix must be a pandas.DataFrame in the format:
index feature_1 feature_2 ... feature_N A ... ... ... ... B ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... Each column will be handled as a feature and tested for its significance to the target.
The target vector must be a pandas.Series or numpy.array in the form
index target A ... B ... . ... . ... and must contain all id’s that are in the feature matrix. If y is a numpy.array without index, it is assumed that y has the same order and length than X and the rows correspond to each other.
Examples
>>> from tsfresh.examples import load_robot_execution_failures >>> from tsfresh import extract_features, select_features >>> df, y = load_robot_execution_failures() >>> X_extracted = extract_features(df, column_id='id', column_sort='time') >>> X_selected = select_features(X_extracted, y)
Parameters: - X (pandas.DataFrame) – Feature matrix in the format mentioned before which will be reduced to only the relevant features. It can contain both binary or real-valued features at the same time.
- y (pandas.Series or numpy.ndarray) – Target vector which is needed to test which features are relevant. Can be binary or real-valued.
- test_for_binary_target_binary_feature (str) – Which test to be used for binary target, binary feature (currently unused)
- test_for_binary_target_real_feature (str) – Which test to be used for binary target, real feature
- test_for_real_target_binary_feature (str) – Which test to be used for real target, binary feature (currently unused)
- test_for_real_target_real_feature (str) – Which test to be used for real target, real feature (currently unused)
- fdr_level (float) – The FDR level that should be respected, this is the theoretical expected percentage of irrelevant features among all created features.
- hypotheses_independent (bool) – Can the significance of the features be assumed to be independent? Normally, this should be set to False as the features are never independent (e.g. mean and median)
- n_processes (int) – Number of processes to use during the p-value calculation
- chunksize (int) – Size of the chunks submitted to the worker processes
Returns: The same DataFrame as X, but possibly with reduced number of columns ( = features).
Return type: Raises: ValueErrorwhen the target vector does not fit to the feature matrix.
tsfresh.feature_selection.significance_tests module¶
Contains the methods from the following paper about FRESH [2]
Fresh is based on hypothesis tests that individually check the significance of every generated feature on the target. It makes sure that only features are kept, that are relevant for the regression or classification task at hand. FRESH decide between four settings depending if the features and target are binary or not.
The four functions are named
target_binary_feature_binary_test(): Target and feature are both binarytarget_binary_feature_real_test(): Target is binary and feature realtarget_real_feature_binary_test(): Target is real and the feature is binarytarget_real_feature_real_test(): Target and feature are both real
References
| [2] | Christ, M., Kempa-Liehr, A.W. and Feindt, M. (2016). Distributed and parallel time series feature extraction for industrial big data applications. ArXiv e-prints: 1610.07717 https://arxiv.org/abs/1610.07717 |
-
tsfresh.feature_selection.significance_tests.target_binary_feature_binary_test(x, y)[source]¶ Calculate the feature significance of a binary feature to a binary target as a p-value. Use the two-sided univariate fisher test from
fisher_exact()for this.Parameters: - x (pandas.Series) – the binary feature vector
- y (pandas.Series) – the binary target vector
Returns: the p-value of the feature significance test. Lower p-values indicate a higher feature significance
Return type: Raise: ValueErrorif the target or the feature is not binary.
-
tsfresh.feature_selection.significance_tests.target_binary_feature_real_test(x, y, test)[source]¶ Calculate the feature significance of a real-valued feature to a binary target as a p-value. Use either the Mann-Whitney U or Kolmogorov Smirnov from
mannwhitneyu()orks_2samp()for this.Parameters: - x (pandas.Series) – the real-valued feature vector
- y (pandas.Series) – the binary target vector
- test (str) – The significance test to be used. Either
'mann'for the Mann-Whitney-U test or'smir'for the Kolmogorov-Smirnov test
Returns: the p-value of the feature significance test. Lower p-values indicate a higher feature significance
Return type: Raise: ValueErrorif the target is not binary.
-
tsfresh.feature_selection.significance_tests.target_real_feature_binary_test(x, y)[source]¶ Calculate the feature significance of a binary feature to a real-valued target as a p-value. Use the Kolmogorov-Smirnov test from from
ks_2samp()for this.Parameters: - x (pandas.Series) – the binary feature vector
- y (pandas.Series) – the real-valued target vector
Returns: the p-value of the feature significance test. Lower p-values indicate a higher feature significance.
Return type: Raise: ValueErrorif the feature is not binary.
-
tsfresh.feature_selection.significance_tests.target_real_feature_real_test(x, y)[source]¶ Calculate the feature significance of a real-valued feature to a real-valued target as a p-value. Use Kendall’s tau from
kendalltau()for this.Parameters: - x (pandas.Series) – the real-valued feature vector
- y (pandas.Series) – the real-valued target vector
Returns: the p-value of the feature significance test. Lower p-values indicate a higher feature significance.
Return type:
Module contents¶
The feature_selection module contains feature selection algorithms.
Those methods were suited to pick the best explaining features out of a massive amount of features.
Often the features have to be picked in situations where one has more features than samples.
Traditional feature selection methods can be not suitable for such situations which is why we propose a p-value based
approach that inspects the significance of the features individually to avoid overfitting and spurious correlations.